Scientific software management draft
Showing
- .style/header.tex 1 addition, 0 deletions.style/header.tex
- scientific_software_management/bib.bib 24 additions, 0 deletionsscientific_software_management/bib.bib
- scientific_software_management/compile.sh 6 additions, 0 deletionsscientific_software_management/compile.sh
- scientific_software_management/img/eb.png 0 additions, 0 deletionsscientific_software_management/img/eb.png
- scientific_software_management/img/gentoo.png 0 additions, 0 deletionsscientific_software_management/img/gentoo.png
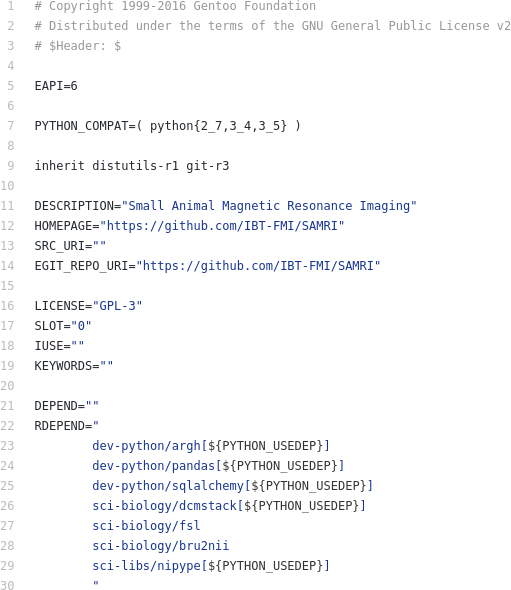
- scientific_software_management/img/ng_mi.png 0 additions, 0 deletionsscientific_software_management/img/ng_mi.png
- scientific_software_management/img/pm.xcf 0 additions, 0 deletionsscientific_software_management/img/pm.xcf
- scientific_software_management/img/pm_d.png 0 additions, 0 deletionsscientific_software_management/img/pm_d.png
- scientific_software_management/img/pm_g.png 0 additions, 0 deletionsscientific_software_management/img/pm_g.png
- scientific_software_management/img/python.png 0 additions, 0 deletionsscientific_software_management/img/python.png
- scientific_software_management/img/sp.png 0 additions, 0 deletionsscientific_software_management/img/sp.png
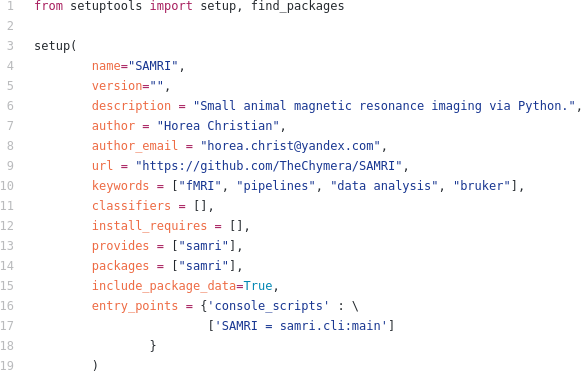
- scientific_software_management/slides.tex 312 additions, 0 deletionsscientific_software_management/slides.tex
scientific_software_management/bib.bib
0 → 100644
scientific_software_management/compile.sh
0 → 100755
scientific_software_management/img/eb.png
0 → 100644
77.7 KiB
107 KiB
scientific_software_management/img/ng_mi.png
0 → 100644
3.6 MiB
scientific_software_management/img/pm.xcf
0 → 100644
File added
scientific_software_management/img/pm_d.png
0 → 100644
16.3 KiB
scientific_software_management/img/pm_g.png
0 → 100644
17 KiB
71.9 KiB
scientific_software_management/img/sp.png
0 → 100644
54.3 KiB
scientific_software_management/slides.tex
0 → 100644